
Setegn Worku Alemu
Quantitative Geneticist






I am a quantitative geneticist specializing in advanced statistical methods for genetic analysis.
I have extensive experience in linear mixed models, Bayesian statistics, and developing professional Shiny applications for genetic data analysis.
Genomics-driven methane reduction in sheep breeding.
Incorporating genomics, RNA, and microbiome data to enhance prediction accuracy for methane emissions in sheep, contributing to climate change mitigation efforts.

Proficient in applying and developing advanced statistical methods for genetic analysis, including linear mixed models, Bayesian approaches, and machine learning techniques in genetics.
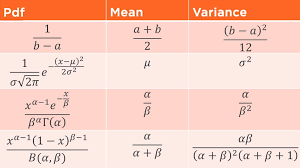
Experienced in developing efficient algorithms and software for large-scale genetic data analysis, with a focus on R, Python, Julia, and JavaScript for high-performance computing in genetics.


Integrates functional genomic information to enhance genomic prediction accuracy.
Analyzes and visualizes LD patterns in complex crossbred populations.
Implements genomic selection models specifically tailored for broiler breeding programs.
Comprehensive dashboard for analyzing and visualizing data from the African Chicken Genetic Gains project.
Incorporates climate data into genetic analyses to study gene-environment interactions.